Examples / Adaptive sampling / gmm
This file is a complete demo of the capability of the gmm function from the CODES toolbox.
Contents
Documentation
The documentation for the gmm function can be found here.
Set rng
Set random number generator seed:
rng(0)
Simple example
Define a 2D reliability problem with 4 failure domains:
f=@CODES.test.inverse_2D; [X,Y]=meshgrid(linspace(-5,5,100)); Z=reshape(f([X(:) Y(:)]),100,100); figure('Position',[200 200 500 500]) contour(X,Y,Z,[0 0],'k')
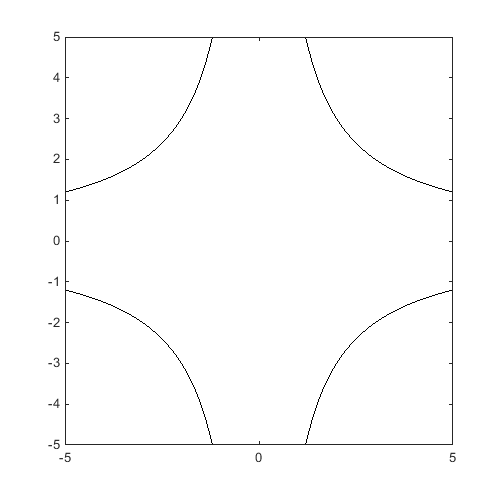
Obtain a DOE:
x=CODES.sampling.cvt(30,2,'lb',[-5 -5],'ub',[5 5],'region',@(x)5-pdist2(x,[0 0])); y=f(x);
Sum of square change at iteration 1: 3.572e-03 Sum of square change at iteration 2: 1.587e-03 Sum of square change at iteration 3: 8.792e-04; Predicted relative change : 2.528e-01 Sum of square change at iteration 4: 5.334e-04; Predicted relative change : 2.163e-01 Sum of square change at iteration 5: 3.657e-04; Predicted relative change : 1.472e-01 Sum of square change at iteration 6: 2.899e-04; Predicted relative change : 6.599e-02 Sum of square change at iteration 7: 2.295e-04; Predicted relative change : 5.380e-02 Sum of square change at iteration 8: 2.055e-04; Predicted relative change : 2.675e-02 Sum of square change at iteration 9: 1.756e-04; Predicted relative change : 1.477e-02 Sum of square change at iteration 10: 1.511e-04; Predicted relative change : 2.727e-03 Sum of square change at iteration 11: 1.251e-04; Predicted relative change : 1.180e-03 Sum of square change at iteration 12: 1.091e-04; Predicted relative change : 5.083e-04 Sum of square change at iteration 13: 9.756e-05; Predicted relative change : 2.180e-04
Derive and check log JPDF and gradient (normal distribution):
logjpdf=@(x)sum(-0.5*x.^2-0.5*log(2*pi),2); % equivalent to logjpdf=@(x)sum(log(normpdf(x)),2); dlogjpdf=@(x)-x; test_rng=@(N)normrnd(0,1,[N 2]); % normal random sampler for optimization starting points x_grad=[0.1 0.1;0.3 0.5;0.8 0.3;0.5 0.5]; grad=dlogjpdf(x_grad); grad_fd=CODES.common.grad_fd(logjpdf,x_grad); disp([['Analytical gradients :';... ' ';... ' Finite differences :';... ' '] num2str([grad';grad_fd'],'%1.3f ')])
Analytical gradients :-0.100 -0.300 -0.800 -0.500
-0.100 -0.500 -0.300 -0.500
Finite differences :-0.100 -0.300 -0.800 -0.500
-0.100 -0.500 -0.300 -0.500
Train an SVM, then find and plot a generalized "max-min" sample:
svm=CODES.fit.svm(x,y); x_gmm=CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf); figure('Position',[200 200 500 500]) contour(X,Y,Z,[0 0],'r') hold on svm.isoplot('lb',[-5 -5],'ub',[5 5],'prev_leg',{'True LS'}) plot(x_gmm(1),x_gmm(2),'ms') axis square
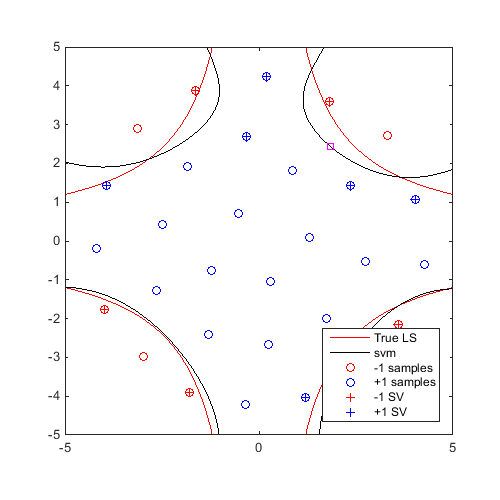
Comparison of Computational Time for: Parallel and Serial; With and Without Derivative; and Matlab and CODES Multistart:
Start a parallel pool:
gcp;
Compare compuational time of different scenarios to compute 10 generalized "max-min" samples
CODES.common.disp_box('Using Matlab MultiStart') tic; svm=CODES.fit.svm(x,y); CODES.sampling.gmm(svm,logjpdf,test_rng,'nb',10,'MultiStart','MATLAB'); disp(['Serial without derivative : ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y,'UseParallel',true); CODES.sampling.gmm(svm,logjpdf,test_rng,'nb',10,'MultiStart','MATLAB'); disp(['Parallel without derivative: ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y); CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf,'nb',10,'MultiStart','MATLAB'); disp(['Serial with derivative: ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y,'UseParallel',true); CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf,'nb',10,'MultiStart','MATLAB'); disp(['Parallel with derivative: ' CODES.common.time(toc)]) CODES.common.disp_box('Using CODES MultiStart') tic; svm=CODES.fit.svm(x,y); CODES.sampling.gmm(svm,logjpdf,test_rng,'nb',10,'MultiStart','CODES'); disp(['Serial without derivative : ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y,'UseParallel',true); CODES.sampling.gmm(svm,logjpdf,test_rng,'nb',10,'MultiStart','CODES'); disp(['Parallel without derivative: ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y); CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf,'nb',10,'MultiStart','CODES'); disp(['Serial with derivative: ' CODES.common.time(toc)]) tic; svm=CODES.fit.svm(x,y,'UseParallel',true); CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf,'nb',10,'MultiStart','CODES'); disp(['Parallel with derivative: ' CODES.common.time(toc)])
########################### # Using Matlab MultiStart # ########################### Serial without derivative : 23s Parallel without derivative: 14s Serial with derivative: 26s Parallel with derivative: 15s ########################## # Using CODES MultiStart # ########################## Serial without derivative : 23s Parallel without derivative: 12s Serial with derivative: 25s Parallel with derivative: 12s
Parallel update
On the same example, find and plot 10 generalized "max-min" samples:
f=@CODES.test.inverse_2D; x=CODES.sampling.cvt(30,2,'lb',[-5 -5],'ub',[5 5],'region',@(x)5-pdist2(x,[0 0])); y=f(x); svm=CODES.fit.svm(x,y,'UseParallel',true); x_gmm=CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf,'nb',10); [X,Y]=meshgrid(linspace(-5,5,100)); Z=reshape(f([X(:) Y(:)]),100,100); figure('Position',[200 200 500 500]) contour(X,Y,Z,[0 0],'r') hold on svm.isoplot('lb',[-5 -5],'ub',[5 5],'prev_leg',{'True LS'}) plot(x_gmm(:,1),x_gmm(:,2),'ms') axis square
Sum of square change at iteration 1: 3.572e-03 Sum of square change at iteration 2: 1.587e-03 Sum of square change at iteration 3: 8.792e-04; Predicted relative change : 2.528e-01 Sum of square change at iteration 4: 5.334e-04; Predicted relative change : 2.163e-01 Sum of square change at iteration 5: 3.657e-04; Predicted relative change : 1.472e-01 Sum of square change at iteration 6: 2.899e-04; Predicted relative change : 6.599e-02 Sum of square change at iteration 7: 2.295e-04; Predicted relative change : 5.380e-02 Sum of square change at iteration 8: 2.055e-04; Predicted relative change : 2.675e-02 Sum of square change at iteration 9: 1.756e-04; Predicted relative change : 1.477e-02 Sum of square change at iteration 10: 1.511e-04; Predicted relative change : 2.727e-03 Sum of square change at iteration 11: 1.251e-04; Predicted relative change : 1.180e-03 Sum of square change at iteration 12: 1.091e-04; Predicted relative change : 5.083e-04 Sum of square change at iteration 13: 9.756e-05; Predicted relative change : 2.180e-04
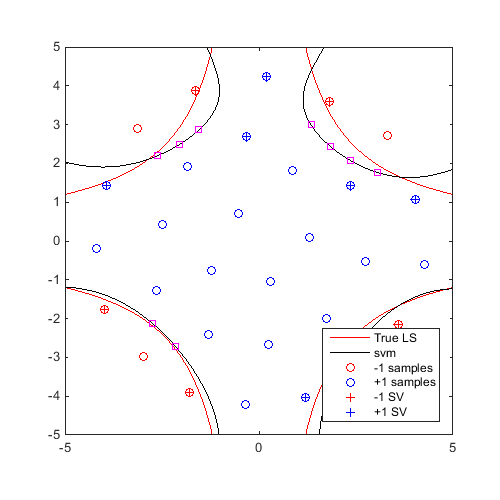
Sequential refinement
On the same example, for 20 iterations, find a "max-min" sample and update the svm:
f=@CODES.test.inverse_2D; x=CODES.sampling.cvt(30,2,'lb',[-5 -5],'ub',[5 5],'region',@(x)5-pdist2(x,[0 0])); y=f(x); svm=CODES.fit.svm(x,y,'UseParallel',true); [X,Y]=meshgrid(linspace(-5,5,100)); Z=reshape(f([X(:) Y(:)]),100,100);
Sum of square change at iteration 1: 3.572e-03 Sum of square change at iteration 2: 1.587e-03 Sum of square change at iteration 3: 8.792e-04; Predicted relative change : 2.528e-01 Sum of square change at iteration 4: 5.334e-04; Predicted relative change : 2.163e-01 Sum of square change at iteration 5: 3.657e-04; Predicted relative change : 1.472e-01 Sum of square change at iteration 6: 2.899e-04; Predicted relative change : 6.599e-02 Sum of square change at iteration 7: 2.295e-04; Predicted relative change : 5.380e-02 Sum of square change at iteration 8: 2.055e-04; Predicted relative change : 2.675e-02 Sum of square change at iteration 9: 1.756e-04; Predicted relative change : 1.477e-02 Sum of square change at iteration 10: 1.511e-04; Predicted relative change : 2.727e-03 Sum of square change at iteration 11: 1.251e-04; Predicted relative change : 1.180e-03 Sum of square change at iteration 12: 1.091e-04; Predicted relative change : 5.083e-04 Sum of square change at iteration 13: 9.756e-05; Predicted relative change : 2.180e-04
Plot first and last iteration:
x_gmm=CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf); svm=svm.add(x_gmm,f(x_gmm)); figure('Position',[200 200 500 500]) contour(X,Y,Z,[0 0],'r') hold on svm.isoplot('lb',[-5 -5],'ub',[5 5],'prev_leg',{'True LS'}) plot(x_gmm(:,1),x_gmm(:,2),'ms') axis square axis square for i=2:20 x_gmm=CODES.sampling.gmm(svm,logjpdf,test_rng,'dlogjpdf',dlogjpdf); svm=svm.add(x_gmm,f(x_gmm)); end figure('Position',[200 200 500 500]) contour(X,Y,Z,[0 0],'r') hold on svm.isoplot('lb',[-5 -5],'ub',[5 5],'prev_leg',{'True LS'}) plot(x_gmm(:,1),x_gmm(:,2),'ms') axis square axis square
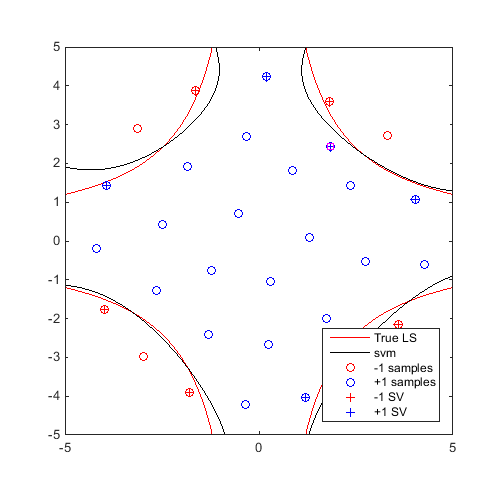
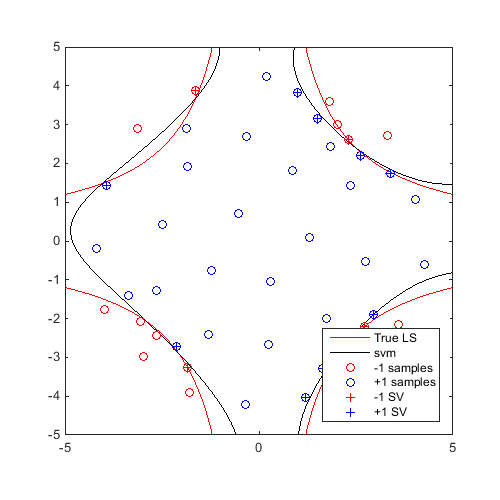
A video of this sequential update can be found here.
Copyright © 2015 Computational Optimal Design of Engineering Systems (CODES) Laboratory. University of Arizona.

|
Computational Optimal Design of Engineering Systems |

|